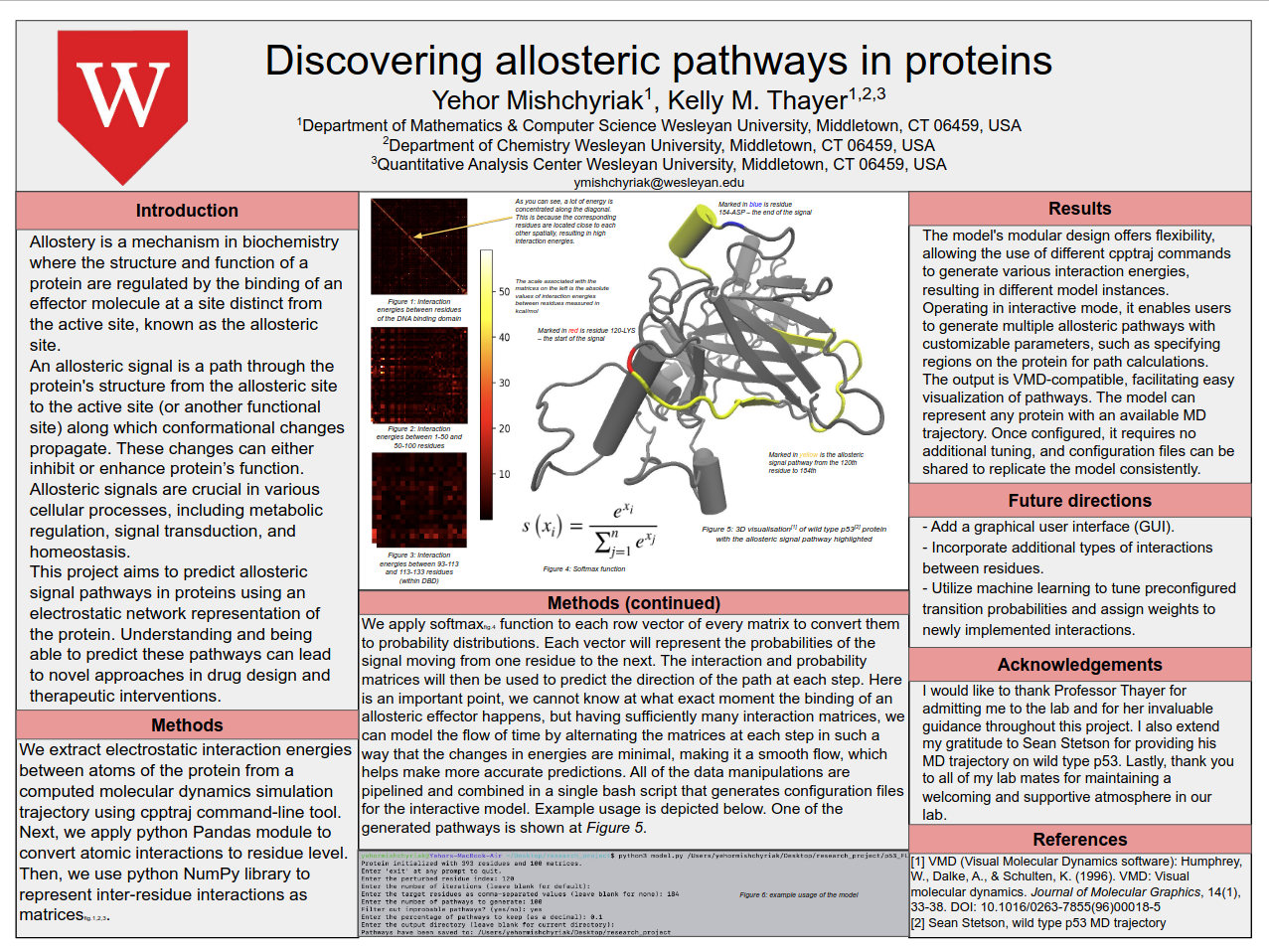
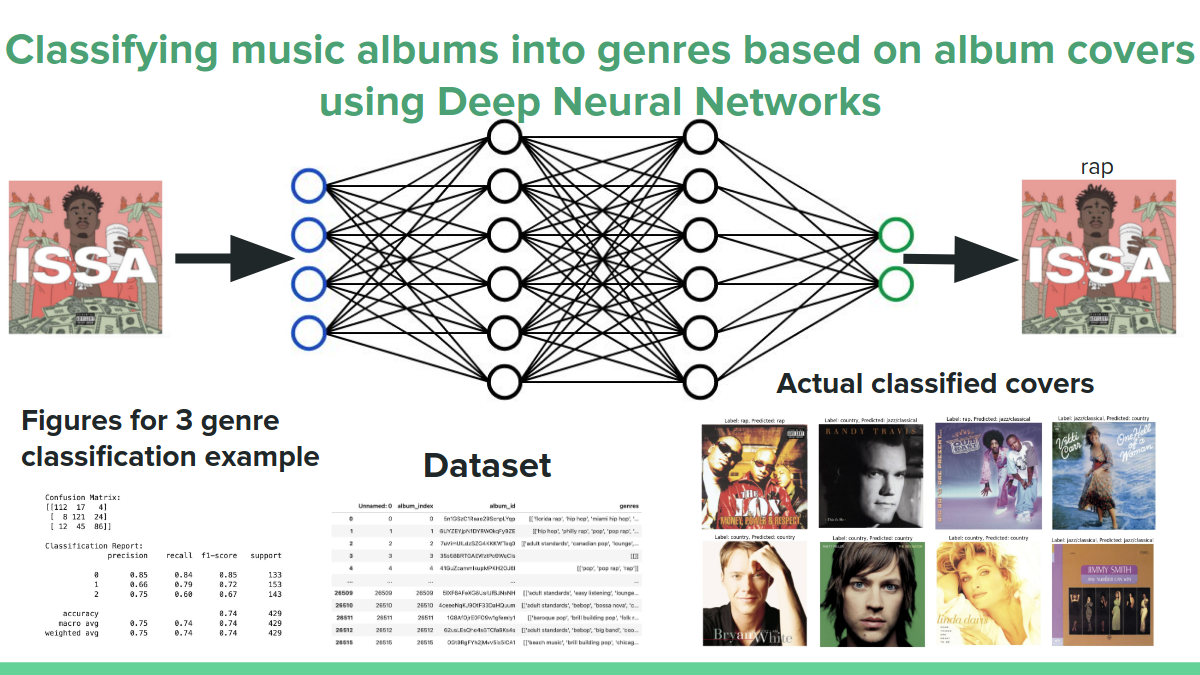
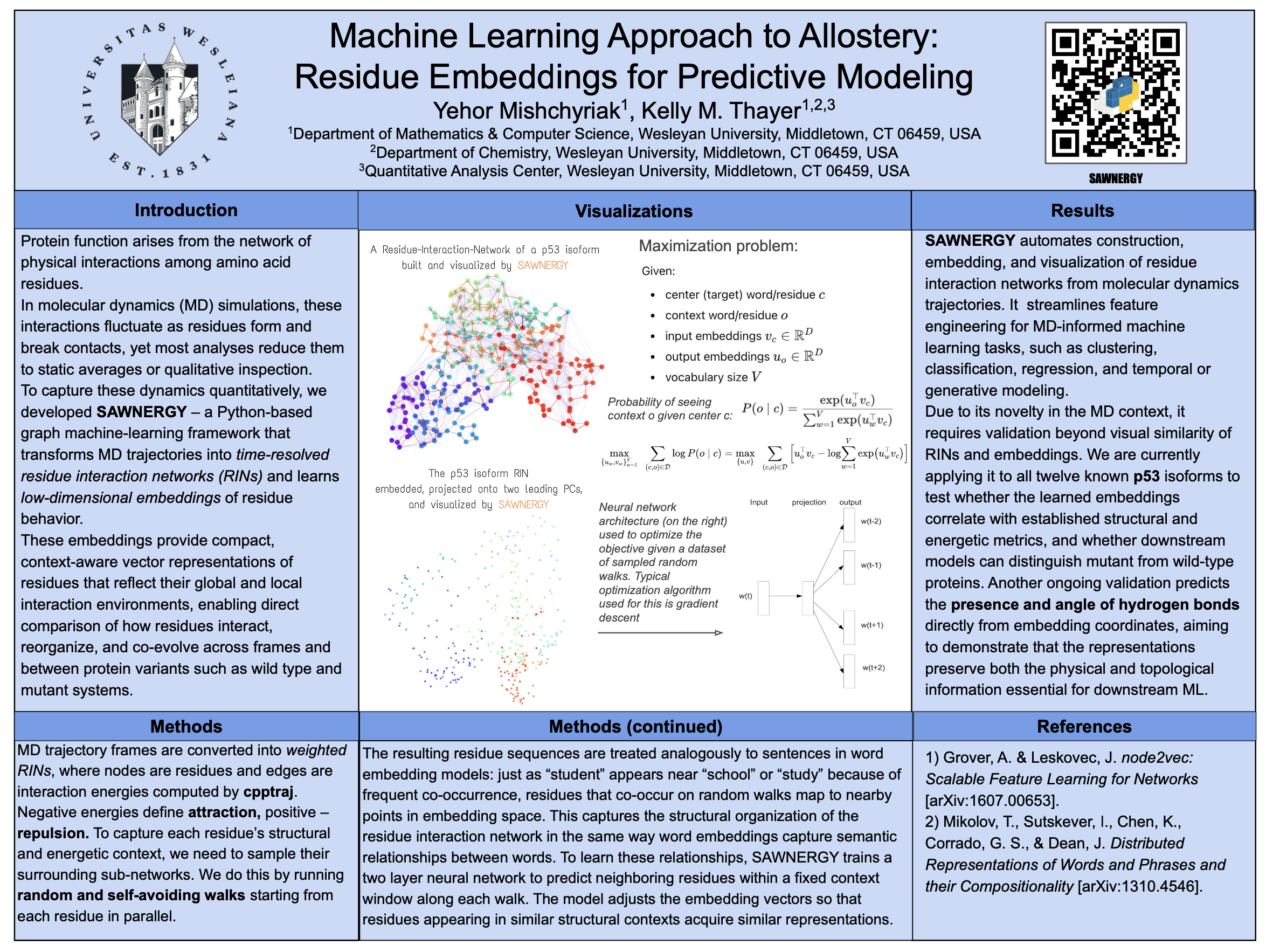
Yehor Mishchyriak
I’m an undergraduate from Ukraine studying Computer Science and Mathematics at Wesleyan University in Middletown, CT. I split my research time between ThayerLab at Wesleyan and BonhamLab at Tufts University School of Medicine. My work sits at the intersection of machine learning, software engineering, and computational biology. At ThayerLab, I develop methods that turn molecular dynamics simulations into machine-learning-ready representations, enabling large-scale analysis of protein allostery. At BonhamLab, I build deep-learning and graph-based models for functional annotation of proteins in the human gut microbiome.