SAWNERGY Documentation

A Python 3.11+ toolkit that converts molecular dynamics (MD) trajectories into residue interaction networks (RINs), samples random and self-avoiding walks (RW/SAW), trains skip-gram embeddings (PureML or PyTorch backends), and visualizes both networks and embeddings. All artifacts are Zarr v3 archives stored as compressed .zip files with rich metadata so every stage can be reproduced.




Requirements
- Python >= 3.11.
cpptraj(AmberTools) discoverable onPATHor viaCPPTRAJ,AMBERHOME/bin, orCONDA_PREFIX/bin.RINBuilderrefuses to run without it.- Default dependencies are installed via
pip install sawnergy; PureML (ym-pure-ml) is bundled. PyTorch is optional but required for themodel_base="torch"embedding backend (GPU if available).
Installation
pip install sawnergy
# If you want the PyTorch backend, install torch separately:
# pip install torch
Small visual example (constructed fully from trajectory and topology files)
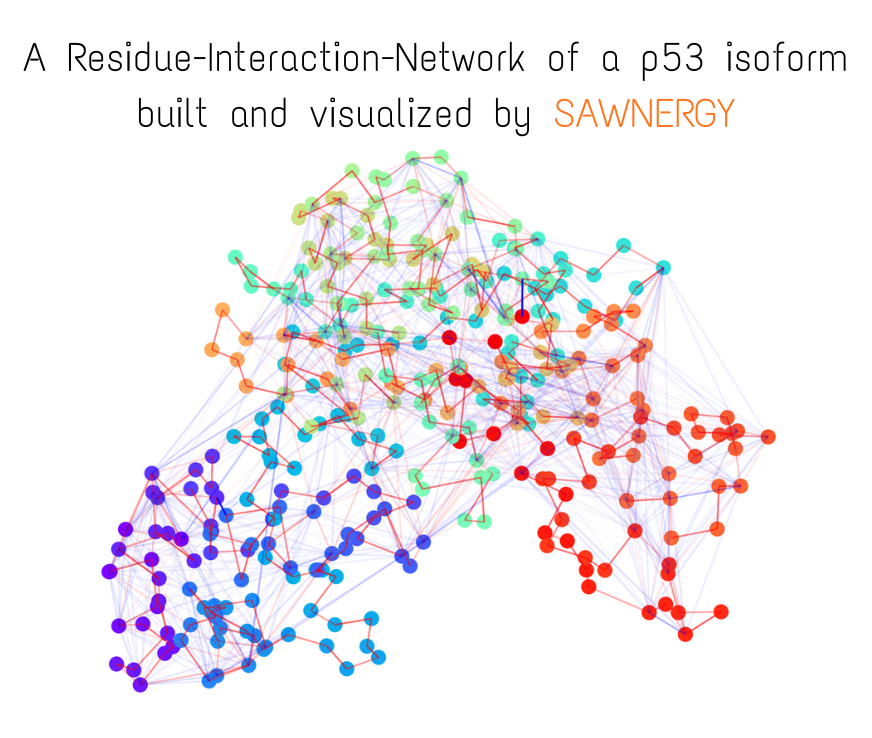
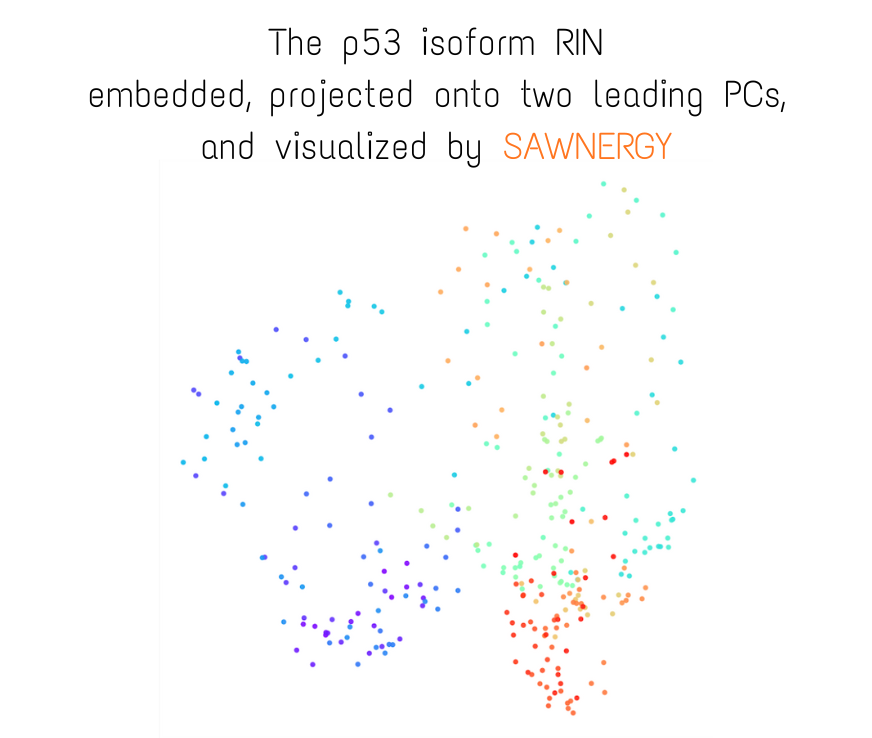
More visual examples:
Animated Temporal Residue Interaction Network of Full Length p53 Protein
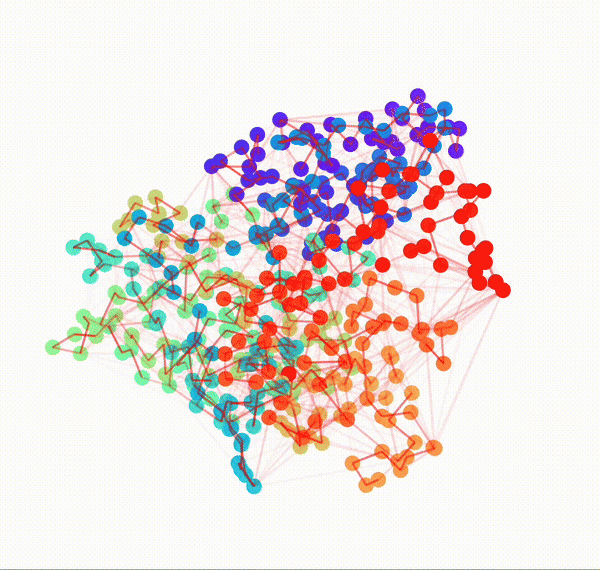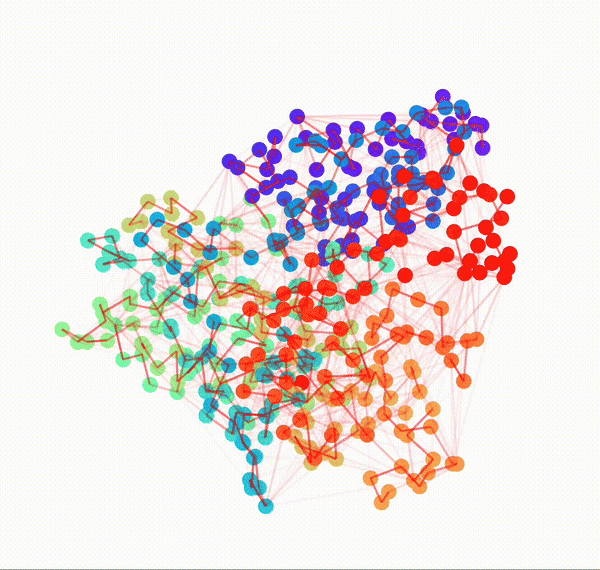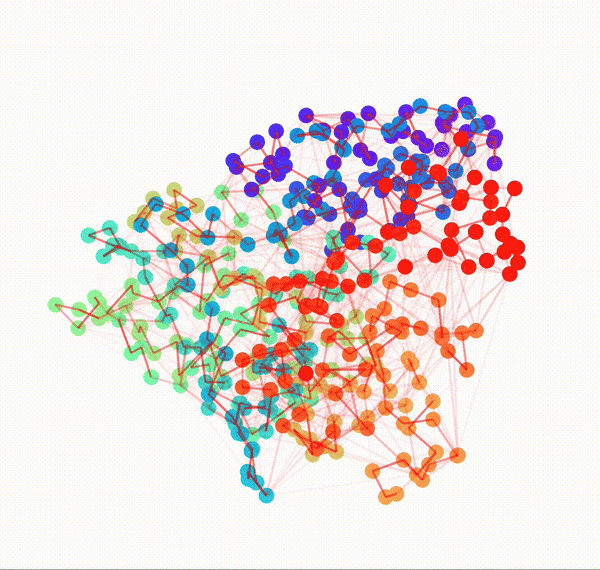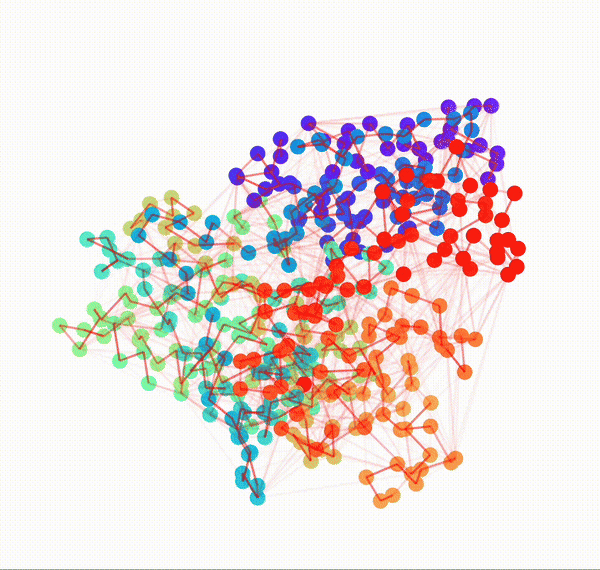
Residue Interaction Network of Full Length p53 Protein (on the right) and its Embedding (on the left)
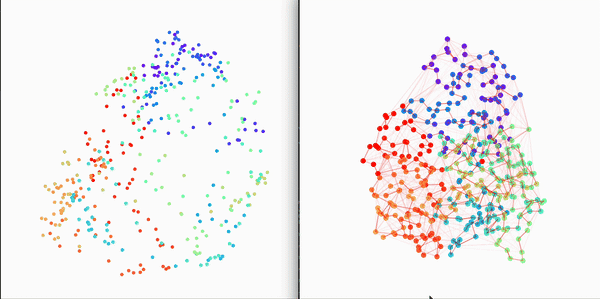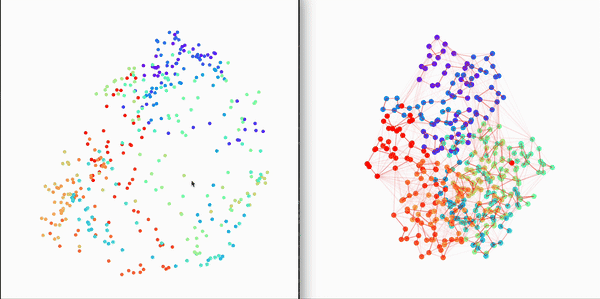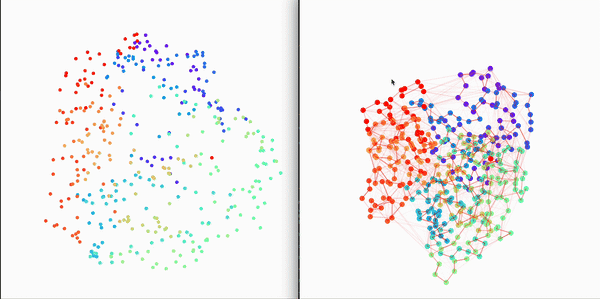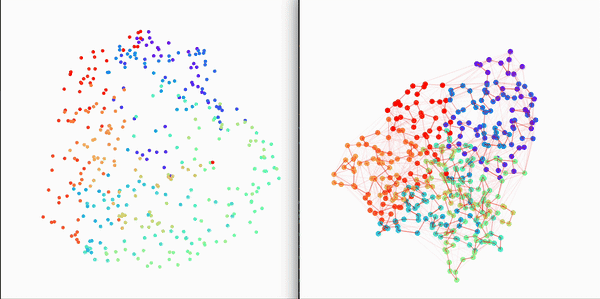
End-to-End Quick Start (make sure cpptraj is discoverable)
New here? This is the shortest possible runnable script. Point it at your own topology/trajectory pair or at the bundled p53 example (see Quick-start MD example below).
from pathlib import Path
import logging
import torch # optional, only when using model_base="torch"
from sawnergy.logging_util import configure_logging
from sawnergy.rin import RINBuilder
from sawnergy.walks import Walker
from sawnergy.embedding import Embedder
configure_logging("./logs", file_level=logging.WARNING, console_level=logging.INFO, force=True)
# 1) Build a Residue Interaction Network archive
rin_path = Path("RIN_demo.zip")
rin_builder = RINBuilder() # auto-locates cpptraj
rin_builder.build_rin(
topology_file="system.prmtop",
trajectory_file="trajectory.nc",
molecule_of_interest=1,
frame_range=(1, 100), # inclusive, 1-based
frame_batch_size=10, # processed in 10-frame batches
prune_low_energies_frac=0.85, # per-row quantile pruning
include_attractive=True,
include_repulsive=False,
output_path=rin_path,
)
# 2) Sample random / self-avoiding walks
walks_path = Path("WALKS_demo.zip")
with Walker(rin_path, seed=123) as walker:
walker.sample_walks(
walk_length=16,
walks_per_node=100,
include_attractive=True,
include_repulsive=False,
time_aware=False,
output_path=walks_path,
in_parallel=False, # required kwarg; set True only under a main-guard
)
# 3) Train per-frame embeddings
embedder = Embedder(walks_path, seed=999)
emb_path = embedder.embed_all(
RIN_type="attr", # "attr" or "repuls"
using="merged", # "RW" | "SAW" | "merged"
num_epochs=20,
negative_sampling=True, # SG (False) or SGNS (True)
num_negative_samples=10,
window_size=5,
device="cuda" if torch.cuda.is_available() else "cpu",
model_base="torch",
kind="in", # stored embedding kind
output_path="EMBEDDINGS_demo.zip",
)
print("Embeddings written to", emb_path)
MD Trajectory + Topology
│
▼
RINBuilder
│ → RIN archive (.zip/.zarr) → Visualizer (display/animate RINs)
▼
Walker
│ → Walks archive (RW/SAW per frame)
▼
Embedder
│ → Embedding archive (frame × vocab × dim)
▼
Downstream ML
Each stage consumes the archive produced by the previous one. Metadata embedded in the archives ensures frame order,
node indexing, and RNG seeds stay consistent across the toolchain.
Biophysical intuition (why these steps exist)
- Residue Interaction Networks treat each amino acid as a node and the non-bonded interaction energy it exchanges with neighbors as a weighted edge. We split energies into attractive (Coulomb + vdW < 0) and repulsive (> 0) channels to respect the underlying physics: attractive edges guide paths along likely contact surfaces; repulsive edges highlight clashes or steric barriers.
- Quantile pruning removes the weakest interactions (default: bottom 85% of each row) so transient noise or solvent “fuzz” does not dominate the walk statistics. We keep the strongest contributors that are most likely to mediate allosteric communication.
- Row-wise L1 normalization converts the energy map into a transition probability matrix. The walker then behaves like a residue-to-residue diffusion process biased by interaction strength, which is a reasonable proxy for signal or force propagation paths through the protein.
- Random walks explore common routes; self-avoiding walks emphasize diverse, non-redundant paths. Time-aware walks allow transitions between frames when transition matrices remain similar, capturing slowly evolving conformations.
- Skip-gram embeddings treat walk paths like sentences: residues that frequently co-occur along walks share similar vectors. This encodes community structure (e.g., domains, loops around a binding pocket) and how it shifts over time.
Quick-start MD example
A minimal dataset is included in example_MD_for_quick_start/ on GitHub to let you run the full SAWNERGY pipeline immediately:
p53_DBD.prmtop(topology),p53_DBD.pdb(reference),p53_DBD.nc(trajectory)- 1 µs production trajectory of the p53 DNA-binding domain, 1000 snapshots saved every 1 ns
- Credits: MD simulation produced by Sean Stetson (ORCID: 0009-0007-9759-5977)
- Intended use: quick-start tutorial for building RINs, sampling walks, and training embeddings without setting up your own MD run
See example_MD_for_quick_start/brief_description.md.
Five-minute “first run” for newcomers
- Install:
pip install sawnergy(andpip install torchif you want GPU training). Confirmcpptraj -hworks in your shell or setCPPTRAJ=/path/to/cpptraj. - Download the p53 quick-start folder and
cdinto the repo root. - Run the quick-start script above with
topology_file="example_MD_for_quick_start/p53_DBD.prmtop"andtrajectory_file="example_MD_for_quick_start/p53_DBD.nc". - Inspect the outputs:
RIN_demo.zip,WALKS_demo.zip, andEMBEDDINGS_demo.zip(all are Zarr-in-zip archives). Trysawnergy.visual.Visualizer("RIN_demo.zip").build_frame(1, show=True)to see the network. - Swap in your own trajectory/topology once you’ve seen the expected behavior.
Archive Layouts (Zarr v3 in .zip)
All archives are Zarr v3 groups and can be opened directly with sawnergy.sawnergy_util.ArrayStorage (read via mode="r"; write/append via mode="a"/"w"). When compressed as .zip, they are read-only; create/append uses the .zarr directory form or a temporary store prior to compression.
| Archive | Core datasets (name → shape, dtype) | Key root attrs |
|---|---|---|
| RIN | ATTRACTIVE_transitions (T, N, N) float32 (opt) • REPULSIVE_transitions (T, N, N) float32 (opt) • ATTRACTIVE_energies (T, N, N) float32 (opt; pre-normalized) • REPULSIVE_energies (T, N, N) float32 (opt) • COM (T, N, 3) float32 |
com_name="COM" • molecule_of_interest • frame_range (tuple or None) • frame_batch_size • prune_low_energies_frac • attractive_transitions_name / repulsive_transitions_name (may be None) • attractive_energies_name / repulsive_energies_name (may be None) • time_created |
| Walks | ATTRACTIVE_RWs (T, N·num_RWs, L+1) uint16 (opt) • REPULSIVE_RWs (T, N·num_RWs, L+1) uint16 (opt) • ATTRACTIVE_SAWs (T, N·num_SAWs, L+1) uint16 (opt) • REPULSIVE_SAWs (T, N·num_SAWs, L+1) uint16 (opt) |
seed • num_workers • in_parallel • batch_size_nodes • num_RWs • num_SAWs • node_count • time_stamp_count • walk_length • walks_per_node • dataset name attrs for each channel (may be None) • walks_layout="time_leading_3d" • time_created |
| Embeddings | FRAME_EMBEDDINGS (T, N, D) float32 |
frame_embeddings_name • time_stamp_count • node_count • embedding_dim • model_base • embedding_kind (`"in" |
T equals the number of frame batches produced by RINBuilder (i.e., frame_range swept in frame_batch_size steps). Walk node ids are 1-based in storage; embedding training converts them to 0-based internally.
Stage Reference
RINBuilder (sawnergy.rin.RINBuilder)
- Purpose: run
cpptraj, compute per-frame atomic EMAP/VMAP energies, project to residues, split into attractive/repulsive channels, prune, symmetrize, L1-normalize into transition matrices, and write a compressed archive. - Construction:
RINBuilder(cpptraj_path=None)auto-resolvescpptrajby checking an explicit path,CPPTRAJ,PATH,AMBERHOME/bin, thenCONDA_PREFIX/bin(verification viacpptraj -h). build_rin(...)(returns output path asstr):- Required:
topology_file,trajectory_file,molecule_of_interest(e.g.,1for^1mask in CPPTRAJ),frame_range(tuple or None). - Defaults:
frame_batch_size=-1(all selected frames in one batch),prune_low_energies_frac=0.85,keep_prenormalized_energies=True,include_attractive=True,include_repulsive=True,parallel_cpptraj=False,simul_cpptraj_instances=None(→os.cpu_count()),num_matrices_in_compressed_blocks=10,compression_level=3. - Processing per batch:
cpptraj pairwiseover the selected frame range → EMAP + VMAP → summed atomic interaction matrix (float32).- Project to residues with the compositional mask
R = Pᵀ @ A @ P. - Split into channels (negative → attractive magnitude, positive → repulsive).
- Per-row quantile pruning at
q=prune_low_energies_frac(applied independently to both channels). - Zero diagonals, symmetrize
(M + Mᵀ)/2, then row-wise L1 normalize (transition probabilities; breaks symmetry). - Optionally persist pre-normalized energies; always persist normalized transitions for requested channels.
- Compute per-frame residue COMs for the batch and store their batch mean (so
Tequals the number of batches, not raw frames whenframe_batch_size>1).
- Output: compressed
.zip(Zarr v3). Dataset names are recorded in attrs; absent channels are set toNone. Types are float32 for matrices and COMs. - Notes: cpptraj tasks can be threaded (
parallel_cpptraj=True); BLAS is kept single-threaded in that mode to avoid oversubscription.
- Required:
Walker (sawnergy.walks.Walker)
- Purpose: load transition matrices from a RIN archive into shared memory and sample RW/SAW paths (optionally time-aware).
- Construction:
Walker(RIN_path, seed=None); resolves transition dataset names from RIN attrs. Raises if neither channel exists or shapes are not(T,N,N)with matchingN.- Shared memory: matrices live in
SharedNDArray; callclose()(or usewith Walker(...)) to release and unlink segments. - RNG: a master seed is stored; child seeds per batch derive from
numpy.random.SeedSequence.
- Shared memory: matrices live in
- Key methods:
_extract_prob_vector(node, time_stamp, interaction_type)returns the transition row (float) for a node/time, renormalized after any masking.walk(start_node=None, start_time_stamp=None, length, interaction_type, self_avoid=False, time_aware=False, stickiness=None, on_no_options=None)→(length+1,)array of 1-based node ids. Whentime_aware=True,stickiness(probability of staying) andon_no_options("raise"or"loop") are mandatory; time steps are chosen by cosine similarity between transition matrices.- SAW dead-ends: if self-avoidance removes all probability mass for a step, the sampler logs a warning and falls back to an unconstrained RW move instead of raising.
sample_walks(walk_length, walks_per_node, *, saw_frac=0.0, include_attractive=True, include_repulsive=False, time_aware=False, stickiness=None, on_no_options=None, output_path=None, in_parallel, max_parallel_workers=None, compression_level=3, num_walk_matrices_in_compressed_blocks=None)→ path to walks archive.in_parallelis required (process-based viaProcessPoolExecutor); guard withif __name__ == "__main__":whenTrue.- Per-node counts:
num_SAWs = round(walks_per_node * saw_frac),num_RWs = walks_per_node - num_SAWs. - Walk tensors are shaped
(T, total_walks, L+1)with dtypeuint16; time-aware walks live in the layer of their start time. - Metadata includes seeds, worker counts, and dataset names (
ATTRACTIVE_RWs,ATTRACTIVE_SAWs, etc.; missing channels are stored asNone).
- Why walks: they approximate how “signals” or perturbations might diffuse through the contact network. SAWs reduce backtracking, biasing toward alternative corridors; RWs keep a faithful probability flow.
- Recommended usage: keep to regular (time-stationary) walks on the attractive network for now. Mix in a modest SAW fraction (e.g., 0.1–0.3) when you want to encourage exploration of farther residues from each node, similar to the DFS/BFS tradeoff in node2vec’s
p/qtuning. - Experimental flag: time-aware walks are provided for research use but are less battle-tested than stationary walks; prefer the stationary API unless you specifically need temporal hopping.
Embedder (sawnergy.embedding.Embedder)
- Purpose: turn walk corpora into skip-gram training pairs and fit embeddings per frame with either PureML (default) or PyTorch backends.
- Construction:
Embedder(WALKS_path, seed=None); loads available RW/SAW datasets and validates shapes against stored metadata. Walks are 1-based in storage and converted to 0-based internally. - Single frame:
embed_frame(frame_id, RIN_type, using, num_epochs, *, negative_sampling=False, window_size=2, num_negative_samples=10, batch_size=1024, in_weights=None, out_weights=None, lr_step_per_batch=False, shuffle_data=True, dimensionality=128, alpha=0.75, device=None, model_base="pureml", model_kwargs=None, kind=("in",), _seed=None)→ list of(embedding, kind)tuples sorted asavg,in,out.RIN_type:"attr"or"repuls".using:"RW","SAW", or"merged"(concatenates available RW and SAW for that channel).- Negative sampling toggles SGNS vs SG. Noise distributions are built only when
negative_sampling=True. - Warm starts:
in_weightsshape(V,D)for all backends.out_weightsshape(V,D)for SGNS;(D,V)for SG (the SG torch/PureML implementations transpose or expect that shape). - Raises if no pairs are produced or walks are missing.
- All frames:
embed_all(..., kind="in", output_path=None, num_matrices_in_compressed_blocks=20, compression_level=3)→ archive path.- Seeds: master seed stored; per-frame seeds derived deterministically and recorded in
per_frame_seeds. - Warm start across frames: each frame initializes from the previous frame’s embeddings (
outtransposed for SG so shapes match). - Output dataset
FRAME_EMBEDDINGSis float32 with shape(T,N,D), whereTis the walk archive’stime_stamp_count. - Why embeddings: residues that share walk context get nearby vectors, revealing communities or binding-site neighborhoods. Comparing embeddings across frames highlights how those communities reorganize over time (possible allosteric shifts).
- Seeds: master seed stored; per-frame seeds derived deterministically and recorded in
Backends:
model_base="pureml"(default): usesSGNS_PureMLorSG_PureML(no biases). Device hint is ignored (PureML is CPU-only).model_base="torch": usesSGNS_TorchorSG_Torch(no biases). Defaults to CUDA if available unlessdeviceoverrides it.
Visualizers
- RIN Visualizer (
sawnergy.visual.Visualizer):- Loads COM coordinates and optional attractive/repulsive energies from a RIN archive (dataset names resolved via attrs). If an energy channel is absent (
None), that edge layer is skipped. build_frame(frame_id, displayed_nodes="ALL", displayed_pairwise_attraction_for_nodes="DISPLAYED_NODES", displayed_pairwise_repulsion_for_nodes="DISPLAYED_NODES", frac_node_interactions_displayed=0.01, global_interactions_frac=True, global_opacity=True, global_color_saturation=True, node_colors=None, title=None, padding=0.1, spread=1.0, show=False, *, show_node_labels=False, node_label_size=6, attractive_edge_cmap="autumn", repulsive_edge_cmap="winter").- Node selectors are 1-based; validation rejects non-integers before conversion.
- Edge candidates are filtered to the specified nodes, then the heaviest fraction is drawn; color/opacity scaling can be global or restricted to kept edges.
animate_trajectory(start=1, stop=None, step=1, interval_ms=50, loop=False, **build_kwargs)reusesbuild_frame; enforcesshow=Falseduring iteration and finally callsshow()once for a single pass. Negativestepplays backwards.- Backend handling:
ensure_backend(show)picks a GUI backend; ifshow=Truebut no display is available, falls back toAggand emits a warning.
- Loads COM coordinates and optional attractive/repulsive energies from a RIN archive (dataset names resolved via attrs). If an energy channel is absent (
- Embedding Visualizer (
sawnergy.embedding.Visualizer):- Loads
FRAME_EMBEDDINGSfrom an embeddings archive; optionalnormalize_rows=TrueL2-normalizes rows before PCA. build_frame(frame_id, *, node_colors="rainbow", displayed_nodes="ALL", show_node_labels=False, show=False)projects the selected frame to 3D via SVD-based PCA (pads to 3 coordinates ifD<3). Node selectors are 1-based and validated.- Shares color semantics with the RIN visualizer (
node_colorscan be a colormap string, per-node RGBA array shaped(N,3|4), or group tuples).
- Loads
Example code
from sawnergy.visual import Visualizer
v = Visualizer("./RIN_demo.zip")
v.build_frame(1,
node_colors="rainbow",
displayed_nodes="ALL",
displayed_pairwise_attraction_for_nodes="DISPLAYED_NODES",
displayed_pairwise_repulsion_for_nodes="DISPLAYED_NODES",
show_node_labels=True,
show=True
)
Visualizer lazily loads datasets and works even in headless environments (falls back to the Agg backend).
from sawnergy.embedding import Visualizer
viz = Visualizer("./EMBEDDINGS_demo.zip", normalize_rows=True)
viz.build_frame(1, show=True)
Utilities
ArrayStorage: thin wrapper over Zarr v3 groups backed by a.zarrdirectory or read-only.zip. Handles per-block chunk metadata, JSON-safe attrs, block iteration, and compression viacompress(into=..., compression_level)or context-managedcompress_and_cleanup(output_pth, compression_level).writeappends along axis 0;readandblock_iterreturn NumPy arrays (copies). Default chunk length when unset is 10 with a warning.- Root attrs always include
array_chunk_size_in_block,array_shape_in_block, andarray_dtype_in_block.
logging_util.configure_logging(logs_dir, file_level=logging.WARNING, console_level=logging.WARNING, force=False)installs a timed rotating file handler plus console handler. Whenforce=True, existing root handlers are removed first.
Practical Notes
Walker.sample_walksuses process-based parallelism; wrap calls inif __name__ == "__main__":whenin_parallel=True. Shared memory segments are unlinked only by the creating process; callclose()(or use a context manager) to avoid leaks.- Time-aware walks require
stickinessin[0,1]andon_no_optionsset to"raise"or"loop". Time transitions are chosen by cosine similarity between transition matrices, renormalized before sampling. - Transition matrices coming from
RINBuilderare row-normalized probabilities;_step_noderenormalizes again after any avoidance masks to keep probabilities valid. - Frame dimension
Tequals the number of frame batches, not necessarily the raw frame count ifframe_batch_size > 1. - Missing channels are represented by
Nonein attrs; downstream stages skip them gracefully (e.g., repulsive walks/edges/embeddings are absent if never built). - Stability note: time-aware walks remain experimental; stick to stationary walks on the attractive network plus a small SAW fraction when you want deeper-but-not-hopping exploration.
Project Structure
├── sawnergy/
│ ├── rin/ # RINBuilder and cpptraj integration helpers
│ ├── walks/ # Walker class and shared-memory utilities
│ ├── embedding/ # Embedder + SG/SGNS backends (PureML / PyTorch)
│ ├── visual/ # Visualizer and palette utilities
│ │
│ ├── logging_util.py
│ └── sawnergy_util.py
│
└── README.md
Minimal API Cheatsheet
- Build RIN:
RINBuilder().build_rin(topology_file, trajectory_file, molecule_of_interest, frame_range, frame_batch_size=-1, prune_low_energies_frac=0.85, keep_prenormalized_energies=True, include_attractive=True, include_repulsive=True, parallel_cpptraj=False, simul_cpptraj_instances=None, num_matrices_in_compressed_blocks=10, compression_level=3) - Walks:
Walker(rin_path, seed=None).sample_walks(walk_length, walks_per_node, saw_frac=0.0, include_attractive=True, include_repulsive=False, time_aware=False, stickiness=None, on_no_options=None, output_path=None, in_parallel=False, max_parallel_workers=None, compression_level=3, num_walk_matrices_in_compressed_blocks=None) - Embeddings:
Embedder(walks_path, seed=None).embed_all(RIN_type, using, num_epochs, negative_sampling=False, window_size=2, num_negative_samples=10, batch_size=1024, lr_step_per_batch=False, shuffle_data=True, dimensionality=128, alpha=0.75, device=None, model_base="pureml", model_kwargs=None, kind="in", output_path=None, num_matrices_in_compressed_blocks=20, compression_level=3) - Visualization:
sawnergy.visual.Visualizer(rin_path, show=False).build_frame(...)or.animate_trajectory(...);sawnergy.embedding.Visualizer(emb_path, normalize_rows=False).build_frame(...).
All functions raise informative ValueError/RuntimeError when inputs are inconsistent (e.g., missing walks, out-of-range frame ids, invalid quantiles). Attributes recorded in each archive are intended to be sufficient to reproduce downstream stages without additional bookkeeping.
